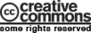Phylogeny tools
Neil Caithness & Milo Thurston (UOXF)
|
KEY RESOURCES |
|---|
| The Oxford Batch Operation Engine |
Setting the scene for integrating services
Batch operations
The integration of external computational tools and services (e.g. identification keys, phylogenetic tools) for data analysis within Scratchpads or any other virtual research environment requires a well designed and documented API (Application Program Interface). An API is a set of protocols and tools for building a software application. A good API makes it easier to develop a program by providing all the building blocks.
A service has been mounted, called OBOE (Oxford Batch Operation Engine). OBOE is the infrastructure that makes it possible to provide external computing services to consumers through Virtual Research Environments (VREs), such as Scratchpads.
The purpose is to run analyses that might normally be performed by a self-contained command-line application and to make them available via the web. The catalogue of services is itself available from the API and, as new services come on-line, the calling VRE will include the new option in the menu of services.
The API design plan...
if you aren't concerned with the technicalities, then skip this section and go to 'Phylogenetic services' below.
Workflow
-
A Scratchpad posts a job request to the Job Scheduler (software program) at Oxford. Authentication is simple as there is currently only one authentic user (i.e. the Scratchpad server at the NHM), but will grow to a small number of such clients when we begin to support external servers.
-
The Job Scheduler enters the job in its Job Table and retrieves data files based on information in the post. The job is sent to the appropriate service (at Oxford or elsewhere). On completion of the job the Job Scheduler updates the Job Table with a job completed status flag and an output URL.
-
The Scratchpad server polls the Job Table until the completed status flag is returned and then fetches the output from the supplied URL.
Implementation
-
Developed with Ruby on Rails with MongoDB as the database. This choice of database is anticipated as allowing more flexibility for dealing with job parameters than a standard relational database.
-
Scratchpad to post data to the Job Scheduler, comprising of job type, user id and a URL for the data files. The system will then get the files. Post to be on port 443 using certificate authentication.
-
Information to be stored in a "jobs" table using a "job" model. This will contain:
- Rails job id
- Calling server id (currently always the NHM Scratchpad server)
- Calling server supplied user id
- Job type
- Location of data file(s) (URL)
- Job status (an integer, pending, sent, processing, finished, error etc.)
-
An array of any parameters required for the job.
-
Whilst the job is running the Scratchpad should poll a URL using their job ID which would find_by_id and return the info. by rendering JSON.
-
Submission to the remote service would presumably have to be by means of a Rake task run from cron, to process any pending jobs and attempt to submit them to the processing service.
-
An implication for the Scratchpad server(s) is that the Scratchpad code would require forms for job submission to be added, and certificates would be needed on the server(s) for authentication against one held at Oxford.
-
Details of the data processing service still undetermined but we hope that it will be able to post queries back (see the separate data_service_post controller above).
Computing platforms
The OBOE system now consists of at least nine machines:
- Unix physical machines (Vibrant, Bonnacon, Cockatrice, Karakal)
- Unix virtual machines (Goblin, Voonith)
- Windows physical machines (Gecko)
- Windows virtual machines (Nyogtha, Syncerus)
We have in principle agreements and a free allocation of startup CPU hours to run services on the OSC clusters and on NGS resources to allow us to quantify demand and establish sustainability costs.
A simple service API is now in place that will allow any authenticated Windows or Unix machine, located anywhere on the internet, to run OBOE services.
Metadata repository
The metadata, including job configuration data, is stored in a repository as an array of replicated MongoDB databases. This is an integral part of the OBOE platform.
We will at some stage, though, review the job metadata storage. This would be most appropriate with the review of user feedback. We anticipate that the metadata will be fed back to the client and stored with the data so that a given task can be run at some future date.
Phylogenetic services
As far as possible existing software for phylogenetic analysis (GARLI, PAUP, TNT, Mr Bayes etc.) will be wrapped for deployment on the various platforms described above.
This task is progressing more slowly than was anticipated. Atttempts to contact the Lattice project (community working on networking systems) and LIRMM (computer science laboratory) have been unsuccesful.
The Lisbeth software for cladistic research is not yet available for Unix platforms, though as OBOE now includes Windows VMs as available platforms, running Lisbeth as a service can be reinvestigated.
 |
The OBOE (Oxford Batch Operation Engine) platform
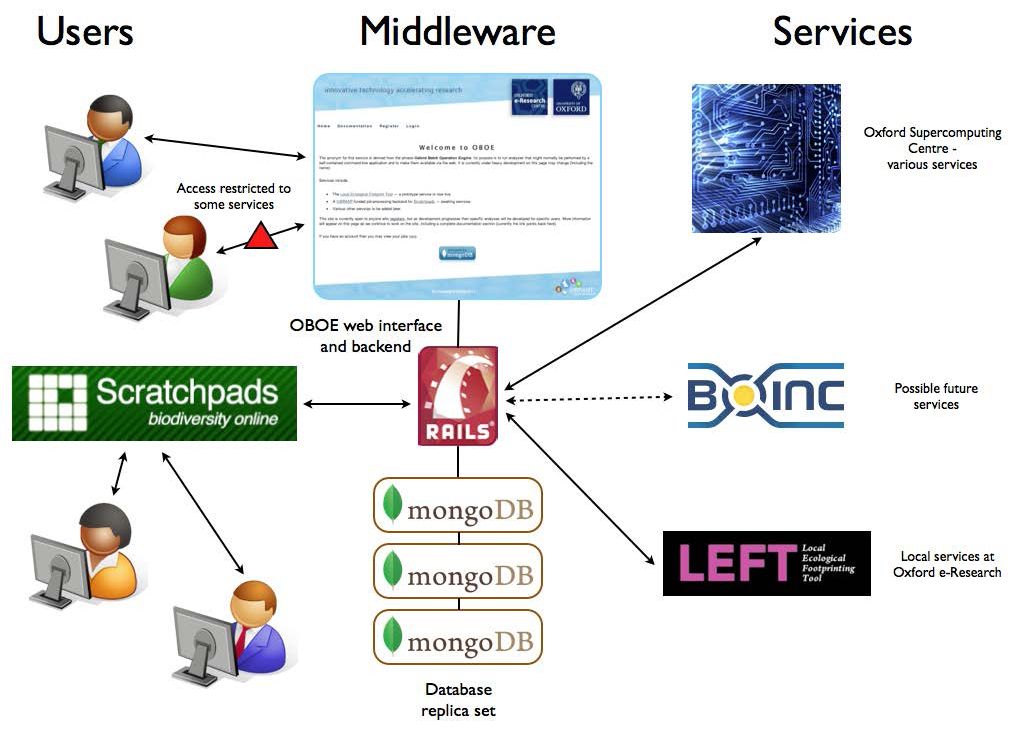
We have been investigating a one-click process from data retrieval (molecular) to alignment and phylogenetic analysis along the lines of LIRMM above. For data retrieval we have experimented with Genbank, for alignment with Clustalw and Muscle, and for phylogenies, Phylip. Automating a seamless process is not straightforward and without risks but we believe it is achievable and potentially useful within the OBOE framework.
We are currently collaborating with the BioVeL project to implement workflows through the workflow management system Taverna.